Recently I was facing an optimization problem: I needed to produce a largish data frame of cash flow projections. This turned out to be a rather slow operation.
The main issue being that in my use case it was not feasible to pre-allocate the final structure ahead, so I had to make do with building the output piece by piece via repeated rbind calls.
The base::rbind() function is okay when called once or twice per script, but when used thousands of times in a row it gets rather slow; the root cause being memory management.
This drew me to investigating three alternatives to the base scenario, a study in optimization that I would like to share with you:
-
dplyr::bind_rows()from the authors of mighty{tidyverse} -
data.table::rbindlist()from the alternative realm of{data.table} - spinning up an in memory SQLite database, and running inserts of intermediary results in the SQL way
As my original use case was needlessly complicated I am illustrating the three approaches on a simplified use case of pasting together 5000 copies of the popular mtcars dataset.
For measuring the time elapsed I am using the simple, yet elegant, {tictoc} package.
tictoc::tic()
for (i in 1:5000) {
if (i == 1) {
res_base <- mtcars
} else {
res_base <- rbind(res_base, mtcars)
}
}
tictoc::toc()## 311.291 sec elapsedThe base approach was just that: basic. It took five minutes to run, and established a baseline for evaluating performance of the more sophisticated approaches.
tictoc::tic()
for (i in 1:5000) {
if (i == 1) {
res_dplyr <- mtcars
} else {
res_dplyr <- dplyr::bind_rows(res_dplyr, mtcars)
}
}
tictoc::toc()## 269.352 sec elapsedThe dplyr approach is very similar to base – both in syntax and speed (which was somewhat unfortunate, as I was expecting a better performance).
Unless you are running a closed tidyverse shop I am not recommending this solution.
tictoc::tic()
for (i in 1:5000) {
if (i == 1) {
res_dtable <- mtcars
} else {
res_dtable <- data.table::rbindlist(list(res_dtable, mtcars))
}
}
tictoc::toc()## 23.228 sec elapsedThe data.table asks for a slightly modified syntax, as the rbidlist() function requires a list of data frames as argument. It also was, in line with the package reputation, blazingly fast.
This approach is probably the best from a general perspective.
tictoc::tic()
con <- DBI::dbConnect(RSQLite::SQLite(), ":memory:")
for (i in 1:5000) {
if (i == 1) DBI::dbCreateTable(con, "mtcars", mtcars)
DBI::dbAppendTable(con, "mtcars", mtcars)
}
res_sqlite <- DBI::dbReadTable(con, "mtcars")
DBI::dbDisconnect(con)
tictoc::toc()## 22.036 sec elapsedThe final alternative considered was a relational database. These are normally not associated with fast speed, but the concept of repeated insert was familiar from my SQL background and seemed appropriate for the use case at hand. As the volume of data was not an issue from available memory constraint perspective the database was created in memory.
I was pleasantly surprised that the humble database was able to keep up with the best that R world has to offer (i.e. data.table) and handily beat the dplyr solution.
On the other hand the benefits of an in memory database approach, when compared to the data.table solution, are only slight and probably worth it only to the hardest core of SQL fans.
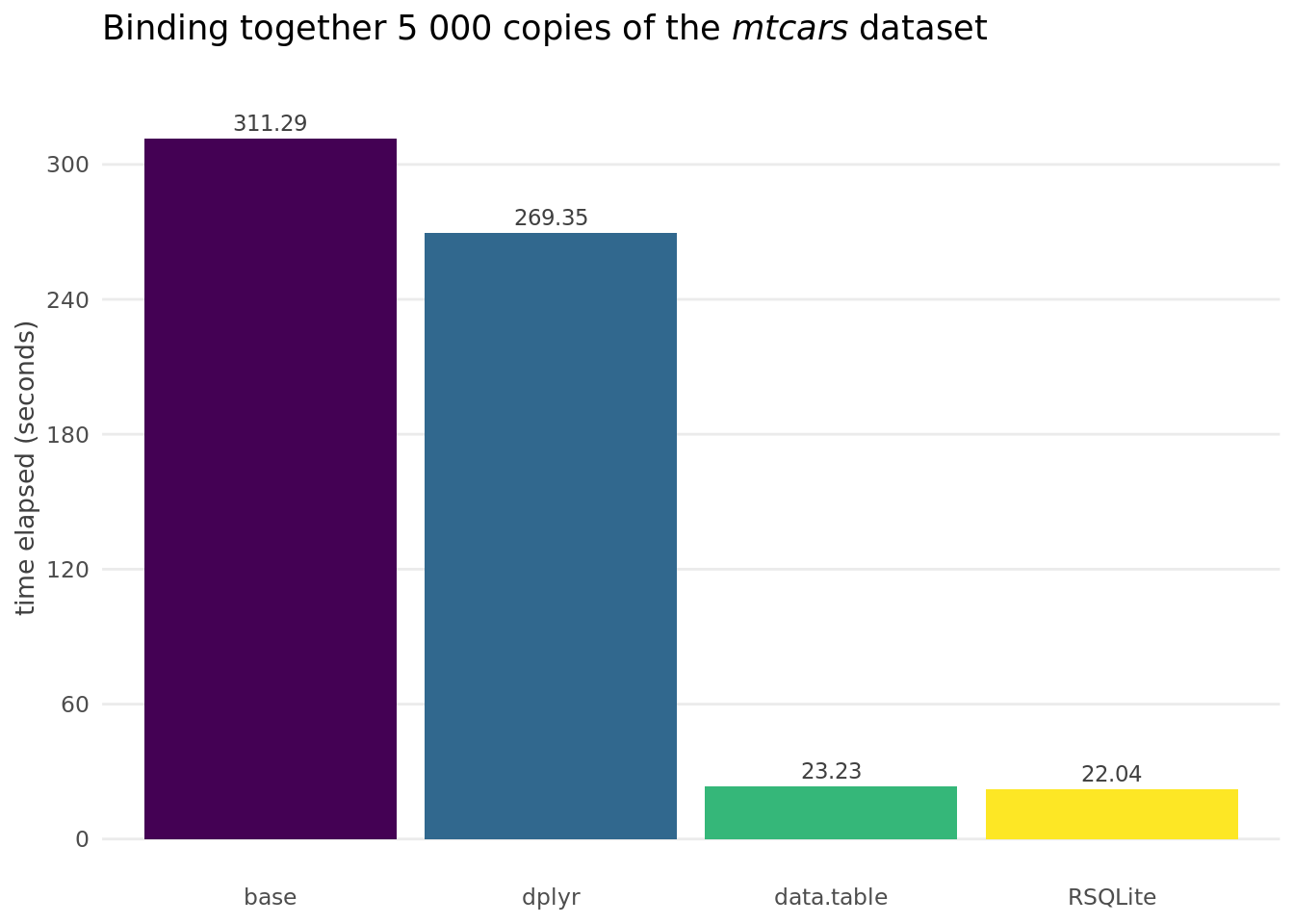
And finally, because an image is worth a thousand of words, a graphical overview of the times elapsed.